Posters
73rd conference of the Hellenic Society of Biochemistry and Molecular Biology, Athens, 1-3 December 2023.
Eleni Poulou-Sidiropoulou1, Charalampos N. Bompas1, Martina Samiotaki2, Alexios Vlamis-Gardikas1
1 Department of Chemistry, Universi ty of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
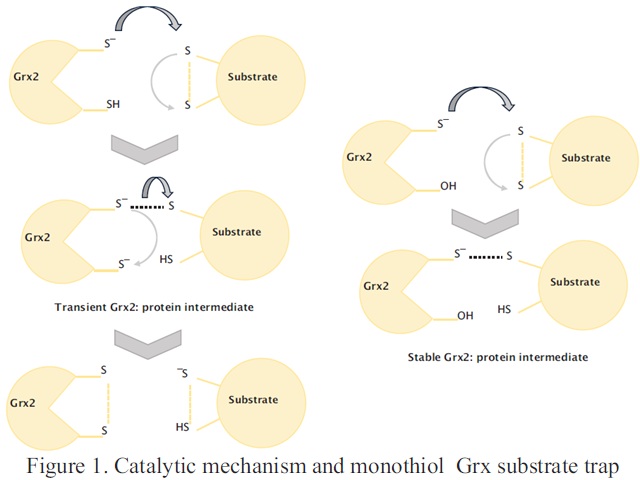
Introduction
Glutaredoxin 2 (Grx2) contributes more that 80 % of glutathione (GSH) mediated redox activity in cellular extracts and protects cells from general oxidative damage (formation of carbonyls1,2). The specific substrates of Grx2 however, are unknown. To this aim, immobilized monothiol/athiol Grx2 mutants were used in affinity chromatography with cellular lysates from Escherichia coli (E. coli). The athiol Grx2 (Grx2 C9S C12S) served as a bait for proteins interacting in a non-thiol manner while the monothiol Grx2 C12S was used to trap dithiol substrates of Grx2 (Figure 1, right column). In addition, lysates from E. coli null mutants for grxB, encoding Grx2, were compared to those of the wild type. All proteomic analyses were performed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations.
Methodology
Overexpression and purification of E. coli Grx2 C9S C12S and Grx2 C12S mutants. Each protein (approximately 6 mg) was immobilized on Affi-Gel 10 beads in chromatographic columns. E. coli cells were grown in LB-medium and collected during both exponential and stationary phases. Cell lysates were prepared and chromatographed through columns with of the immobilized Grx2mutants. Empty resin served as control. Bound proteins were eluted with salt (KCl, step gradients), CH3COOH/HCOOH and finally DTT. The eluted proteins were analyzed by LC-MS/MS. All cromatographies were performed in triplicates. Furthermore, E. coli wild type strain and the null mutant for grxB were grown and harvested in their exponential phase of growth. Their total proteins were analyzed by LC-MS/MS.
Charalampos N. Bompas1, Eleni Poulou Sidiropoulou1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1*
1 Department of Chemistry, Universi ty of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
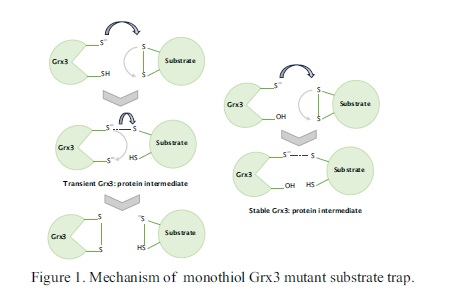
Introduction
Glutaredoxin 3 (Grx3) catalyses thiol-disulfide exchange reactions between protein substrates and glutathione (GSH)1,2 but its biological role is unknown. Its only biologically relevant activity is the inefficient in vitro reduction of ribonucleotide reductase 1a3. To investigate its function, affinity chromatography was employed for Escherichia coli cell lysates through columns with immobilized monothiol Grx3 mutant. Possible substrates of dithiol Grx3 are expected to interact with the monothiol Grx3 species (Figure 1, right column). In addition, lysates from E. coli null mutants of the grxC gene encoding Grx3, were compared to those of the wild type. All proteomic analyses were performed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations.
Methodology
TheE.coli Grx3 C14S C65Y mutant wasoverexpressed and purified. Approximately 6 mg of Grx3 were immobilized per mL of Affi-Gel 15 beads and placed in chromatographic columns. E. coli cells were grown in LB-medium and harvested at the exponential and stationary growth phases to provide a pool of possible interacting partners. Chromatography of the lysate supernatants followed under increasing salt (KCl), acid (CH3COOH/HCOOH) and reductive (DTT) conditions. All experiments were performed thrice. Eluants were analyzed with LC-MS/MS. Proteomic comparisons at the exponential phase of growth were also performed between the wild-type and the grxC null mutant. All protein fractions were analyzed by LC-MS/MS.
International Conference of Traditional Medicine, Immunology, Biochemistry and Food Technology. Cognition Conferences, Zurich, 25-26 November 2024.
Eleni Poulou-Sidiropoulou1, Charalampos N. Bompas1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1*.
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.

Introduction
Glutaredoxin 3 (Grx3) catalyses thiol-disulfide exchange reactions between protein substrates and glutathione (GSH)1,2 but its biological role is unknown. Its only biologically relevant activity is the inefficient in vitro reduction of ribonucleotide reductase 1a3. To investigate its function, affinity chromatography was employed for Escherichia coli cell lysates through columns with immobilized monothiol Grx3 mutant. Possible substrates of dithiol Grx3 are expected to interact with the monothiol Grx3 species (Figure 1, right column). In addition, lysates from E. coli null mutants of the grxC gene encoding Grx3, were compared to those of the wild type. All proteomic analyses were performed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations.
Methodology
The E.coli Grx3 C14S C65Y (monothiol) mutant was overexpressed, purified and immobilized on Affi-Gel 15 resin. Approximately 6 mg of Grx3 were immobilized per mL of gel beads. E. coli cells were grown in LB-medium and harvested at the exponential and stationary phases of growth. Affinity chromatography of selected cell lysates started with increasing salt (KCl) concentrations to be followed by acidic (CH3COOH/HCOOH) and finally reducing (DTT) conditions. All experiments were performed thrice. Eluants were analysed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations. In addition, whole proteome comparisons were performed between the wild-type and the grxC null mutant for the exponential and stationary phases of growth.
Charalampos N. Bompas1, Eleni Poulou Sidiropoulou1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1*
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
Introduction
Glutaredoxin 3 (Grx3) catalyzes thiol-disulfide exchange reactions between protein substrates and glutathione (GSH)1,2 but its only biologically relevant activity is the inefficient in vitro reduction of ribonucleotide reductase 1a3. To investigate the greater role of Grx3 for the cell, we used affinity columns with immobilized monothiol mutant Grx3, through which cell lysates from Escherichia coli were chromatographed. These columns are expected to trap dithiol substrates of the wild-type Grx3. Further analysis of the interactions between Grxs and identified protein partners was performed by protein docking using the Prism webserver.
Methodology
The E.coli Grx3 C14S C65Y (monothiol) mutant was overexpressed, purified and immobilized on Affi-Gel 15 resin. Approximately 6 mg of Grx3 were immobilized per mL of gel beads. E. coli cells were grown in LB-medium and harvested at the exponential and stationary phases of growth. Affinity chromatography of selected cell lysates started with increasing salt (KCl) concentrations to be followed by acidic (CH3COOH/HCOOH) and finally reducing (DTT) conditions. All experiments were performed thrice. Eluants were analysed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations. In addition, whole proteome comparisons were performed between the wild-type and the grxC null mutant for the exponential and stationary phases of growth.
74th conference of the Hellenic Society of Biochemistry and Molecular Biology, Thessaloniki, 13-15 December 2024.
Charalampos N. Bompas1, Eleni Poulou Sidiropoulou1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1*
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
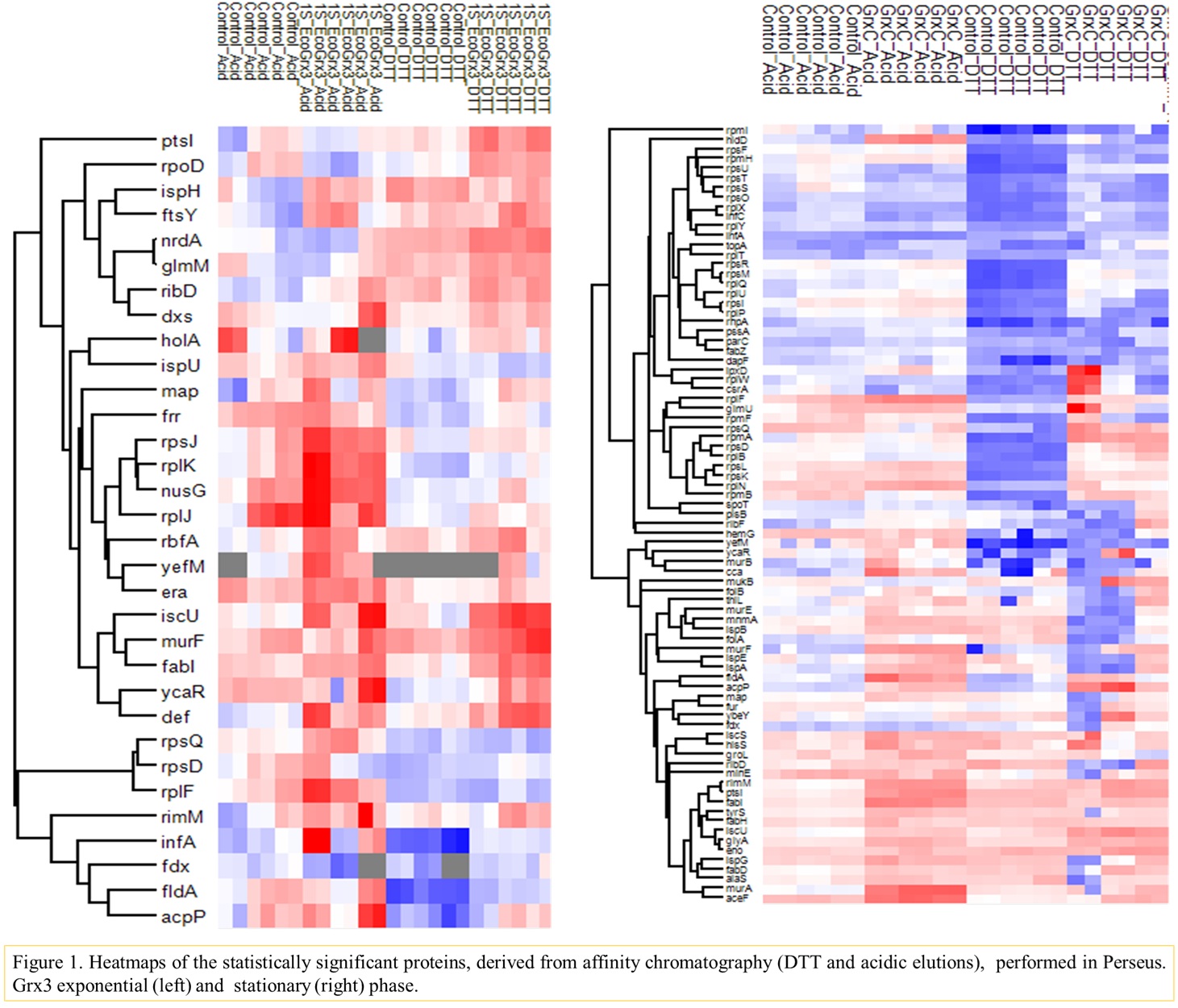
Introduction
Grx2 (encoded by the grxB gene) representing up to 1 % of total soluble protein in the stationary phase of growth, contributes to the protection of cells against oxidative stress induced by H2O2 1. Grx3 (grxC gene) with 0.4 % of total soluble protein may reduce ribonucleotide reductase in vitro. Both Grx2 and 3 participate in thiol-disulfide exchange but their biological role remains unknown. Proteins essential for survival of the Gram-negative pathogen Escherichia coli, may represent novel targets for multitargeting antibiotics2. We have shown that both glutaredoxins (monothiol trapping mechanism3) may interact with essential for survival proteins, whose levels were also altered in null mutants for grxB and grxC. In this work, we examined the interactions of the two glutaredoxins and the essential
Methodology
The protein ligands of E. coli Grx2 and Grx3 were identified by affinity chromatography experiments using as bits monothiol grxs. Cellular lysates corresponded to cells grown to LB medium (exponential and stationary phase). Cell lysates were prepared and chromatographed through columns with of the immobilized Grx2 and Grx3 mutants. Columns with uncoupled resin served as control. Bound proteins were eluted with salt (KCl, step gradients), CH3COOH/HCOOH pH 2,1 and finally DTT. All experiments were performed in triplicates. Furthermore, the whole proteomes of E. coli wild type and the null mutant for grxB and grxC were compared for cells grown in LB (exponential and stationary growth phase). All proteomic analyses were performed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) followed by bioinformatics and gene ontology evaluations.
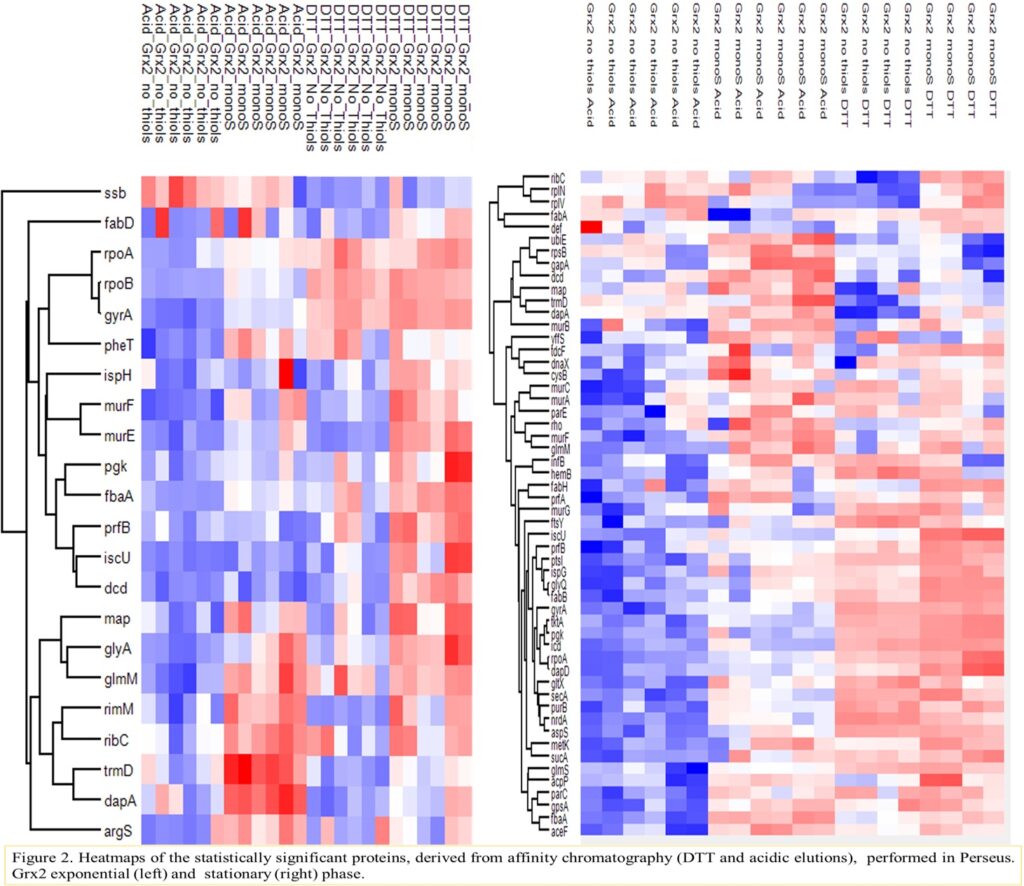
75th conference of the Hellenic Society of Biochemistry and Molecular Biology, Athens, 5-7 December 2025.
Katerina Pegiou1, HaralamposTzoupis1, Rouni Georgia2,Martina Samiotaki2, and Alexios Vlamis-Gardikas1*
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.

Introduction
The glutaredoxin (Grx) and thioredoxin systems catalyze the reversible reduction of disulfides in the cytosol of all organisms including viruses. Glutaredoxin 1 (Grx1) from Escherichia coli (E. coli) is a small (85 aminoacids, 9,7 kDa) cytosolic oxidoreductase that catalyzes glutathione (GSH)–dependent thiol–disulfide exchange reactions. Three more Grxs in the E. coli cytosol, along with Grx1 maintain redox homeostasis. However, Grx1 was initially discovered, and remains, the major electron donor of ribonucleotide reductase, the key enzyme for the de novo biosynthesis of deoxyribonucleotides [1]. All dithiol Grxs have an active site of the type CxxC (C11PYC14 for Grx1) to react via a transient covalent disulfide bond with their substrates [2]. Although this redox activity is well established, the specific covalent and non-covalent interaction partners of Grx1 have not been systematically analyzed. Our study aimed at investigating the largely ignored non-covalent interactions of Grx1. The approach used was affinity chromatography of cell extracts through columns with immobilized Grx1, reduced or oxidized. Our findings combined with in silico analysis suggest that the formation of a simple disulfide in the active site of the enzyme may modify its binding affinity and specificity to its protein
Methodology
In a first step, wild type Grx1 (oxidized or reduced) was used as a bait in affinity chromatography. Cellular lysates from E. coli were prepared and chromatographed under oxidizing (selenite) or reducing (DTT/GSH) conditions through the Grx1-containing affinity columns. Unsubstituted Affi-Gel 15 resin was used as a control. Bound proteins were eluted sequentially by increasing concentrations of KCl, followed by acidic elutions (CH₃COOH/HCOOH, pH 2.1) and finally reduction by DTT (5 mM). All chromatographies (bait and controls) were performed in triplicates. Collected fractions were precipitated with sodium deoxycholate and TCA, washed with methanol, dried, and resuspended in Tris-SDS. The concentrated eluates from each condition were identified by LC-MS/MS and subjected to bioinformatic processing and functional enrichment evaluation. In a second step, the herein identified protein ligands along with all other known protein ligands from UniProt were subjected to in silico docking (Prism) and the binding parameters of the formed complexes were determined by Prodigy. The contacts of the surface residues of reduced and oxidized Grx1 in complexes (by Prism) were detected by Biovia. A percentage representation of all contacts was made after considering all possible contacts of each amino acid of Grx1 with those of the selected protein ligands.
Christos Karamanos1, Εleni Poulou-Sidiropoulou1, Charalampos N. Bompas1, Haralampos Tzoupis1, Theodore Tselios1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
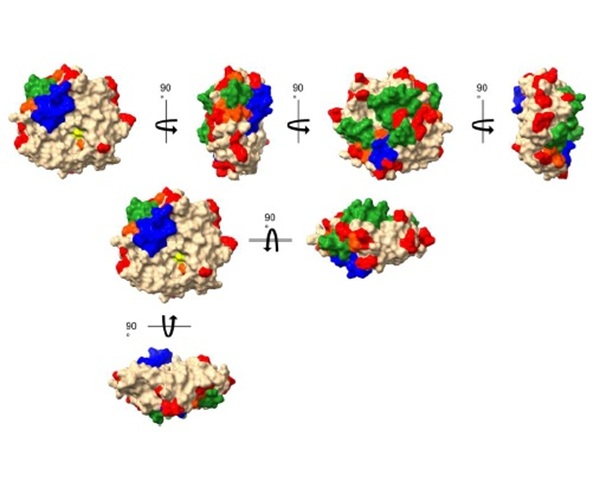
Introduction
The thioredoxin (Trx) and glutaredoxin (Grx) enzyme systems reduce disulfides of target protein substrates in the cytosols of all living species including viruses. The Grx system of Escherichia coli (E. coli) is composed of EcoGrx1-4, glutathione reductase and NADPH [1,2]. EcoGrx2 is an atypical, rather large Grx (215 amino acids, 24,3 kDa) that comprises up to 1 % of total soluble protein in the stationary phase of growth and with strong general antioxidant properties. Its functions however have remained largely unknown [1], until work from our lab that used affinity chromatography and mass spectrometry, highlighted the protein interactome of EcoGrx2. The present work applied in silico approaches to pinpoint the critical amino acid residues (contact hot spots) and greater surfaces for the interactions of EcoGrx2 with its protein ligands.
Methodology
First, PDB files were extract from the Uniprot protein database (https://www.uniprot.org). Prior to molecular docking, heteroatoms and non-protein residues were removed. Protein complexes along with their predicted interfaces and docking scores were created using the web tool Prism [3]. Dissociation constants and Gibbs free energy bindings were calculated using the web tool Prodigy. All complexes (Prism) were analyzed by Biovia to identify the contact residues of EcoGrx2 with its ligands. For each amino acid in each complex, the number and type of interaction was recorded, followed by statistical analysis of its participation in contact interfaces.Τhe most statistically significant residues by Biovia (“contact hot spots” [4]) were compared to the thermodynamic hot spots identified by the web tool Proton (http://proton.tools.ibg.edu.tr). Energy network analysis (https://bioinf.iiit.ac.in/NAPS) was applied on the contact hot spots to highlight potential interfaces for the interactions of EcoGrx2 with its protein ligands.
Christos Karamanos1, Εleni Poulou-Sidiropoulou1, Charalampos N. Bompas1, Haralampos Tzoupis1, Theodore Tselios1, Martina Samiotaki2, and Alexios Vlamis-Gardikas1
1Department of Chemistry, Univer sity of Patras, 26504 Rion, Patras, Greece.
2Institute for Bioinnovation, Biomedical Sciences Research Center “Alexander Fleming”, 16672, Vari, Attica, Greece.
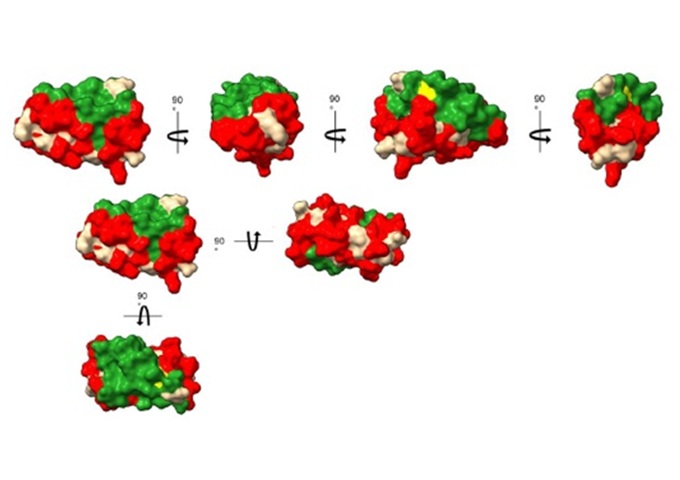
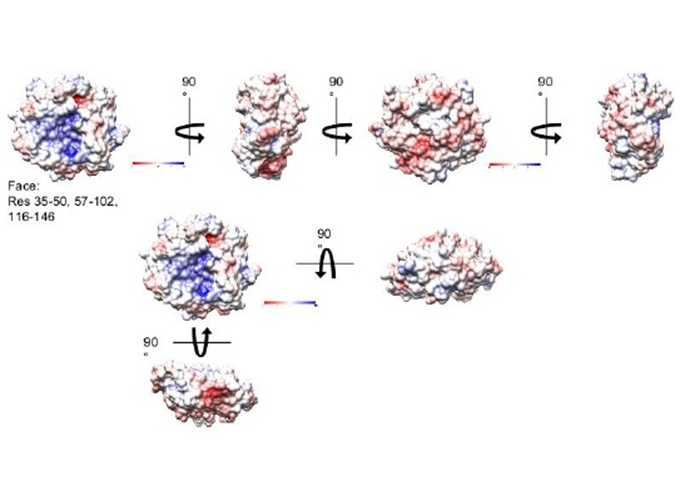
Introduction
The intracellular environment of all cells is kept reduced by the glutaredoxin (Grx) and thioredoxin systems [1]. Escherichia coli (E. coli) has four Grxs (Grx1-4). Glutaredoxin 3 from E. coli (EcoGrx3, 83 amino acids, 9,12 kDa) was discovered in viable null mutants for the two efficient reductors of ribonucleotide reductase Ia (RRIa), thioredoxin 1 and Grx1. RRIa is essential enzyme for the synthesis of deoxyribonucleotides, the precursors of DNA. EcoGrx3 may amount up to 0,5 % of total soluble protein but can barely reduce RRIa, implying that its function(s) may not be confined to the described weak reduction of RRIa [1,2]. Using affinity chromatography, we have previously identified the protein interactome of EcoGrx3. The present work applied in silico approaches to pinpoint the critical amino acid residues (contact hot spots) and greater potential surfaces for interactions of EcoGrx3 with its protein ligands.
Methodology
First, PDB files were extracted from the Uniprot protein database and subjected to molecular docking using the web tool Prism which also gave the predicted interfaces and the respective docking scores [3]. Dissociation constants (KDs) and Gibbs free energy bindings were calculated with the web tool Prodigy. The detection of amino acid contacts and types of interactions was performed by Biovia. The number with which each amino acid of EcoGrx3 participated in the different complexes was used to calculate its relative percentage in forming complexes.Τhis approach gave the statistically significant contact residues (“contact hot spots”) [4]. The predicted thermodynamic hot spots were derived from the webtool Proton. Energy network analysis (https://bioinf.iiit.ac.in/NAPS) was applied on the contact hot spots to give the proposed interfaces of interactions

Attachments
-
 1. Identification of putative interactors of Escherichia coli Glutaredoxin 2.
1. Identification of putative interactors of Escherichia coli Glutaredoxin 2.
-
 2 Interactome identification of Escherichia coli Glutaredoxin 3.
2 Interactome identification of Escherichia coli Glutaredoxin 3.
-
 3. The Putative Interactome of Escherichia coli Glutaredoxin 2 as revealed by Affinity Chromatography
3. The Putative Interactome of Escherichia coli Glutaredoxin 2 as revealed by Affinity Chromatography
-
 4. The protein interactome of Escherichia coli Glutaredoxin 3 Expands its Possible Cellular Functions
4. The protein interactome of Escherichia coli Glutaredoxin 3 Expands its Possible Cellular Functions
-
 5. Escherichia coli glutaredoxins 2 and 3 interact with proteins essential for bacterial survival.
5. Escherichia coli glutaredoxins 2 and 3 interact with proteins essential for bacterial survival.
-
 6. Implications from the updated interactome of Glutaredoxin 1from Escherichia coli
6. Implications from the updated interactome of Glutaredoxin 1from Escherichia coli
-
 7. Mapping the surface areas of Escherichia coli glutaredoxin 2 used for its interaction with protein ligands
7. Mapping the surface areas of Escherichia coli glutaredoxin 2 used for its interaction with protein ligands
-
 8. Escherichia coli glutaredoxin 3: mapping the areas of the molecule interacting with its protein ligands
8. Escherichia coli glutaredoxin 3: mapping the areas of the molecule interacting with its protein ligands

